Flow cytometry#
This use case walks through a sequence of steps that walks through managing a lake of .fcs files as a Flow cytometry lakehouse.
You will
start by ingesting a single file and seed a versioned dataset
append a new batch to the dataset and create a new version
look at an overview of ingested data
query the data and turn it into analytical insights to share back with the team
Setup#
!lamin init --storage ./test-facs --schema bionty
Show code cell output
✅ saved: User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-09-27 19:03:28)
✅ saved: Storage(id='VvPocT7z', root='/home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs', type='local', updated_at=2023-09-27 19:03:28, created_by_id='DzTjkKse')
💡 loaded instance: testuser1/test-facs
💡 did not register local instance on hub (if you want, call `lamin register`)
import lamindb as ln
import lnschema_bionty as lb
import readfcs
lb.settings.species = "human"
💡 loaded instance: testuser1/test-facs (lamindb 0.54.2)
ln.track()
💡 notebook imports: lamindb==0.54.2 lnschema_bionty==0.31.2 pytometry==0.1.4 readfcs==1.1.6 scanpy==1.9.5
💡 Transform(id='OWuTtS4SAponz8', name='Flow cytometry', short_name='facs', version='0', type=notebook, updated_at=2023-09-27 19:03:32, created_by_id='DzTjkKse')
💡 Run(id='6LxzHJKBOJu5s56VPocZ', run_at=2023-09-27 19:03:32, transform_id='OWuTtS4SAponz8', created_by_id='DzTjkKse')
Ingest a first file#
Access  #
#
We start with a flow cytometry file from Alpert et al., Nat. Med. (2019).
Calling the following function downloads the file and pre-populates a few relevant registries:
ln.dev.datasets.file_fcs_alpert19(populate_registries=True)
PosixPath('Alpert19.fcs')
We use readfcs to read the raw fcs file into memory and create an AnnData object:
adata = readfcs.read("Alpert19.fcs")
adata
AnnData object with n_obs × n_vars = 166537 × 40
var: 'n', 'channel', 'marker', '$PnB', '$PnE', '$PnR'
uns: 'meta'
Transform: normalize  #
#
In this use case, we’d like to ingest & store curated data, and hence, we split signal and normalize using the pytometry package.
import pytometry as pm
2023-09-27 19:03:35,878:INFO - Failed to extract font properties from /usr/share/fonts/truetype/noto/NotoColorEmoji.ttf: In FT2Font: Can not load face (unknown file format; error code 0x2)
2023-09-27 19:03:36,045:INFO - generated new fontManager
pm.pp.split_signal(adata, var_key="channel")
'area' is not in adata.var['signal_type']. Return all.
pm.tl.normalize_arcsinh(adata, cofactor=150)
Validate: cell markers  #
#
First, we validate features in .var using CellMarker:
validated = lb.CellMarker.validate(adata.var.index)
❗ 13 terms (32.50%) are not validated for name: Time, Cell_length, Dead, (Ba138)Dd, Bead, CD19, CD4, IgD, CD11b, CD14, CCR6, CCR7, PD-1
We see that many features aren’t validated because they’re not standardized.
Hence, let’s standardize feature names & validate again:
adata.var.index = lb.CellMarker.standardize(adata.var.index)
validated = lb.CellMarker.validate(adata.var.index)
❗ 5 terms (12.50%) are not validated for name: Time, Cell_length, Dead, (Ba138)Dd, Bead
The remaining non-validated features don’t appear to be cell markers but rather metadata features.
Let’s move them into adata.obs:
adata.obs = adata[:, ~validated].to_df()
adata = adata[:, validated].copy()
Now we have a clean panel of 35 validated cell markers:
validated = lb.CellMarker.validate(adata.var.index)
assert all(validated) # all markers are validated
Register: metadata  #
#
Next, let’s register the metadata features we moved to .obs.
For this, we create one feature record for each column in the .obs dataframe:
features = ln.Feature.from_df(adata.obs)
ln.save(features)
We use the Experimental Factor Ontology through Bionty to create a “FACS” label for the dataset:
lb.ExperimentalFactor.bionty().search("FACS").head(2) # search the public ontology
| ontology_id | definition | synonyms | parents | molecule | instrument | measurement | __ratio__ | |
|---|---|---|---|---|---|---|---|---|
| name | ||||||||
| fluorescence-activated cell sorting | EFO:0009108 | A Flow Cytometry Assay That Provides A Method ... | FACS|FAC sorting | [] | None | None | None | 100.0 |
| FACS-seq | EFO:0008735 | Fluorescence-Activated Cell Sorting And Deep S... | None | [EFO:0001457] | RNA assay | None | None | 90.0 |
# import the record from the public ontology and save it to the registry
lb.ExperimentalFactor.from_bionty(ontology_id="EFO:0009108").save()
# show the content of the registry
lb.ExperimentalFactor.filter().df()
| name | ontology_id | abbr | synonyms | description | molecule | instrument | measurement | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||
| lh5Cxy8w | fluorescence-activated cell sorting | EFO:0009108 | None | FACS|FAC sorting | A Flow Cytometry Assay That Provides A Method ... | None | None | None | QAzN | 2023-09-27 19:03:40 | DzTjkKse |
Register: data & annotate with metadata  #
#
modalities = ln.Modality.lookup()
features = ln.Feature.lookup()
efs = lb.ExperimentalFactor.lookup()
species = lb.Species.lookup()
file = ln.File.from_anndata(
adata, description="Alpert19", field=lb.CellMarker.name, modality=modalities.protein
)
... storing '$PnE' as categorical
... storing '$PnR' as categorical
file.save()
Inspect the registered file#
Inspect features on a high level:
file.features
Features:
var: FeatureSet(id='pKU07rLRLFb4YWdJFVmO', n=35, type='number', registry='bionty.CellMarker', hash='ldY9_GmptHLCcT7Nrpgo', updated_at=2023-09-27 19:03:41, modality_id='Pa89rywu', created_by_id='DzTjkKse')
'CD94', 'CD16', 'CD8', 'TCRgd', 'DNA2', 'CD86', 'ICOS', 'PD1', 'CD28', 'CD45RA', ...
obs: FeatureSet(id='AyuadzbYqTo0hOe8d82W', n=5, registry='core.Feature', hash='O72gVchjHlX8pwpZlxqO', updated_at=2023-09-27 19:03:41, modality_id='2CEhNp28', created_by_id='DzTjkKse')
(Ba138)Dd (number)
Cell_length (number)
Bead (number)
Dead (number)
Time (number)
Inspect low-level features in .var:
file.features["var"].df().head()
| name | synonyms | gene_symbol | ncbi_gene_id | uniprotkb_id | species_id | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 0qCmUijBeByY | CD94 | KLRD1 | 3824 | Q13241 | uHJU | RlqM | 2023-09-27 19:03:35 | DzTjkKse | |
| bspnQ0igku6c | CD16 | FCGR3A | 2215 | O75015 | uHJU | RlqM | 2023-09-27 19:03:35 | DzTjkKse | |
| ttBc0Fs01sYk | CD8 | CD8A | 925 | P01732 | uHJU | RlqM | 2023-09-27 19:03:35 | DzTjkKse | |
| ljp5UfCF9HCi | TCRgd | TCRGAMMADELTA|TCRγδ | None | None | None | uHJU | RlqM | 2023-09-27 19:03:35 | DzTjkKse |
| yCyTIVxZkIUz | DNA2 | DNA2 | 1763 | P51530 | uHJU | RlqM | 2023-09-27 19:03:35 | DzTjkKse |
Use auto-complete for marker names:
markers = file.features["var"].lookup()
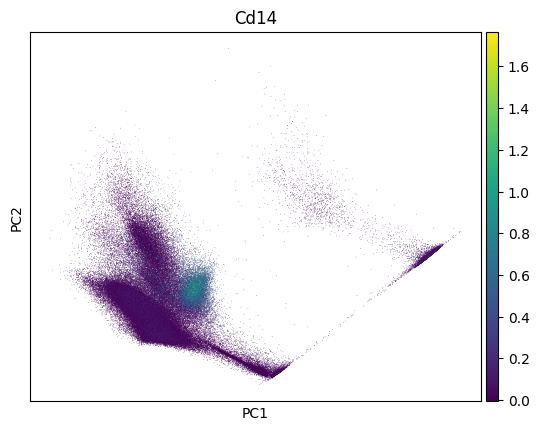
import scanpy as sc
sc.pp.pca(adata)
sc.pl.pca(adata, color=markers.cd14.name)
file.view_flow()

Create a dataset from the file#
dataset = ln.Dataset(file, name="My versioned FACS dataset", version="1")
dataset
Dataset(id='8RZdIbll16NTrAwo7lRL', name='My versioned FACS dataset', version='1', hash='fnzTGHE8BlkiMMIqHLDjyA', transform_id='OWuTtS4SAponz8', run_id='6LxzHJKBOJu5s56VPocZ', file_id='8RZdIbll16NTrAwo7lRL', created_by_id='DzTjkKse')
Let’s inspect the features measured in this dataset which were inherited from the file:
dataset.features
Features:
obs: FeatureSet(id='AyuadzbYqTo0hOe8d82W', n=5, registry='core.Feature', hash='O72gVchjHlX8pwpZlxqO', updated_at=2023-09-27 19:03:41, modality_id='2CEhNp28', created_by_id='DzTjkKse')
(Ba138)Dd (number)
Cell_length (number)
Bead (number)
Dead (number)
Time (number)
This looks all good, hence, let’s save it:
dataset.save()
Annotate by linking FACS & species labels:
dataset.labels.add(efs.fluorescence_activated_cell_sorting, features.assay)
dataset.labels.add(species.human, features.species)
dataset.view_flow()

